Quick navigation: [Introduction] [Interactive Prototype] [How to Help]
[Source Code] [News] [Past Versions]
Try the model in a web browser!
https://nanohub.org/tools/pc4covid19 (v2, April 11, 2020)
Introduction
In March 2020, we assembled an international coalition to develop a comprehensive multiscale simulation framework for SARS-CoV-2 (coronavirus) infections in lung and gut tissues. We aim to understand and test interventions in the coupled dynamics of COVID-19, including:
- Virus spread in tissue
- Virion adhesion to ACE2 receptors on cells
- Endocytosis (active transport into the cell)
- Viral uncoating, replication, and assembly into new virions
- Viral exocytosis (release of completed virions)
- Single-cell responses to infection, including disrupted metabolism, secretion of interferons, and cell death
- Inflammatory responses
- Immune activation and expansion in lymph nodes
- Immune cell infiltration and predation in infected tissue
- Tissue damage, including edema that can lead to acute respiratory distress syndrome (ARDS)
By rapidly creating a multiscale framework, we can ask what if questions that identify vulnerabilities in viral replication and the spread of the infection, and seek approaches to control the immune response to avoid adverse reactions.
To drive this, we have assembled a multidisciplinary team of virologists, mathematical biologists, computer scientists, and industrial pharmacologists, who have all pledged to share data and expertise to proceed much faster as a group than we could alone.
We will share the entire model, scientific documentation, and code as open source, so that the entire community can benefit from this diverse domain expertise and focus on calibrating and validating the model, rather than building and accelerating it. We are sharing our progress with open science principles, including frequent scientific dissemination through interactive web models, open calls for community feedback, and frequent release and update of scientific preprints.
And we are using rapid prototyping: we aim for a 7-14 day release cycle, where each release improved upon the last. Each release includes a well-tested and documented open source code release, an interactive web-hosted version for accelerated scientific communication, public feedback, and an updated preprint.
Preprint: https://github.com/pc4covid19/preprints/blob/master/Rapid_SARS-CoV-2_prototyping-preprint_2.pdf (Update May 8, 2020)
Current interactive prototype
Try the model at https://nanohub.org/tools/pc4covid19.
How can you help?
- Try the model: https://nanohub.org/tools/pc4covid19
- Give feedback:
- Google Form: https://forms.gle/SVUMYWhipSHfX8nS8
- pc4covid19 slack workspace: [invite link]
- Let us know if you can offer data or expertise.
- Read the preprint: https://doi.org/10.1101/2020.04.02.019075
- Spread the word: share this page on twitter
- Keep an eye on this page for improvements!
Source Code
The complete source code and design documents can be found at the pc4covid19 GitHub organization:
https://www.github.com/pc4covid19
News stories
Past versions
v1 prototype (March 25, 2020-April 1, 2020)
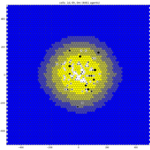
core model: https://github.com/pc4covid19/COVID19/tree/0.1.3
(First release: March 26, 2020; Last update: April 1, 2020)
nanoHUB app: http://dx.doi.org/doi:10.21981/19BB-HM69
(First release: March 26, 2020)
preprint: https://www.biorxiv.org/content/10.1101/2020.04.02.019075v1
v2 prototype (April 2, 2020-present)

core model: https://github.com/pc4covid19/COVID19/releases/tag/0.2.1
(First release: April 9, 2020; Last update: April 10, 2020)
nanoHUB app: http://dx.doi.org/doi:10.21981/J6SP-J909
(First release: April 11, 2020)
preprint: https://github.com/pc4covid19/preprints/blob/master/Rapid_SARS-CoV-2_prototyping-preprint_2.pdf

